Superior HDX-MS
Electrochemical Reduction of Biopharmaceuticals
- Fast and efficient disulfide bond reduction
- Increased sequence coverage in HDX/MS
- Improved characterization of the hinge region of antibodies, cystine knots and other cystine-rich proteins
- Not affecting back-exchange
- Electrons instead of harsh toxic chemicals
Hydrogen–deuterium exchange (HDX) is used for studying the tertiary structure of proteins, or protein folding. Outer regions of the folded protein undergo easy exchange of Hydrogen (H) by Deuterium (D). In the next steps of the workflow, the proteins are unfolded and the amino acid sequence is elucidated by MS. Amino acids with deuterium are obviously from the outer region of the folded protein. For reliable results it is important that the H/D exchange reaction is stopped (quenched), and the changes are preserved, once the unfolding of proteins and the following steps take place.
The controlled electrochemical reduction of the S-S bonds has been successfully applied for the unfolding of larger proteins such as antibodies, replacing the harsh chemical reduction that often jeopardized the H/D exchange. Using classical reducing agents such as DTT or TCEP are often ineffective under quenching conditions (acidic pH and low temperature). Hence, the use of ROXY EC with its proprietary Titanium based electrodes for the gentle and controlled cleavage of the S-S bond opens new opportunities in HDX/MS workflows, resulting in significant higher sequence coverage.
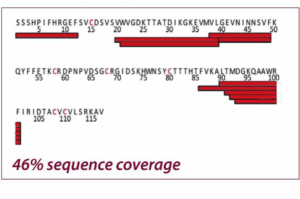
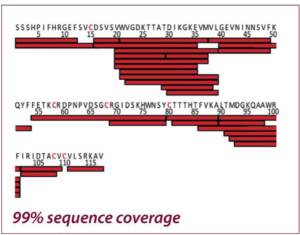
The two figures show the sequence coverage for Nerve Growth Factor-β with chemical and electrochemical (EC) reduction. The integration of the electrochemical flow cell into the HDX/MS workflow resulted in a much higher sequence coverage than chemical reduction (99% vs. 46%) allowing a much more comprehensive characterization of NGF. Adapted from Trajberg E. et al., Anal. Chem.87 (2015) 8880.
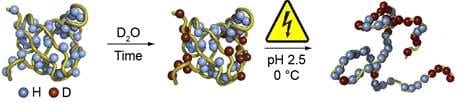
Deuteration, followed by disulfide bond reduction in the workflow of HDX-MS.
Electrochemical Reactors for MS and Synthesis
Environmental Degradation
Lipidomics, Lipid-Oxidation
Proteomics
Application notes
Posters