Amino Acid Trap (AAT) column
- Efficient trapping of amino acids/small peptides
- Easy installation, as a precolumn
- 4 x 50 mm column, 5 µm resin
In compositional analysis of monosaccharides from glycoproteins using HPAEC-PAD, amino acids and small peptides co-elute with the carbohydrates of interest, making proper quantification impossible. These amino acids and small peptides are generated during the acid hydrolysis of glycoproteins. Moreover, amino acids can contaminate the Au electrode surface, which might lead to fouling and loss of response even under PAD conditions. To eliminate the interference of amino acids and to assure accurate quantification of the monosaccharides, the use of an Amino Acid Trap (AAT) column is highly recommended.
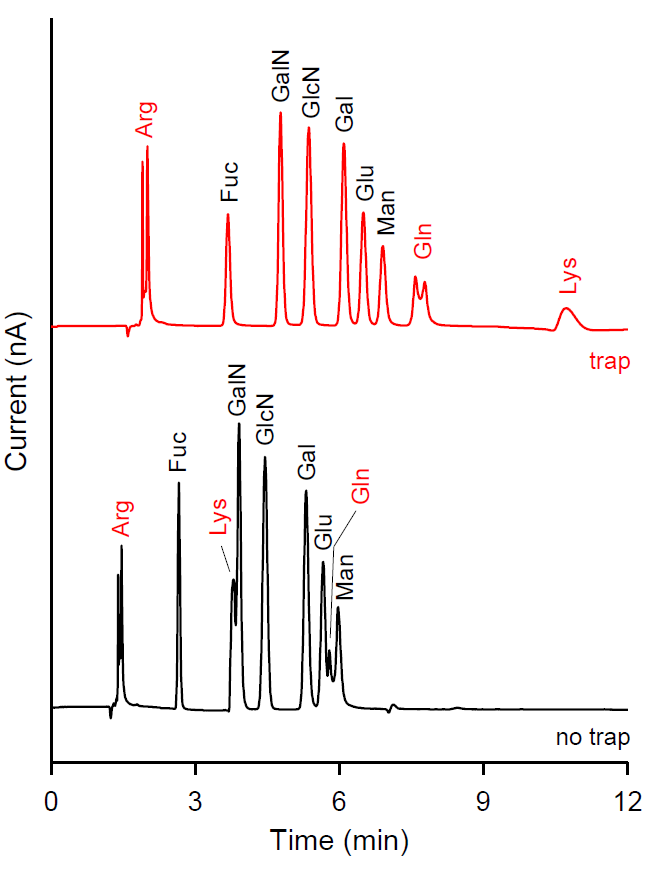
Analysis of monosaccharides from glycoproteins with and without Amino Acid Trap (AAT) column. Interfering peaks such as Glutamine (Gln) and Lysine (Lys) are trapped and elute later during the wash step, see upper trace.